Abstract
Background: Cytomorphology is an essential method to assess disease phenotypes. Recently, promising results of automation, digitalization and machine learning (ML) for this gold standard have been demonstrated. We reported on successful integration of such workflows into our lab routine, including automated scanning of peripheral blood smears and ML-based classification of blood cell images (ASH 2020). Following this pilot project, we are focusing on an equivalent approach for bone marrow.
Aim: To establish a multistep-approach including scan of bone marrow smears and detection/classification of all kinds of bone marrow cell types in healthy individuals and leukemia patients.
Methods: The method includes a pre-scan at 10x magnification for detecting suitable "areas of interest" (AOI) for cytomorphological analysis, a high resolution capture of a predefinable number of AOI at 40x magnification (always using oil) and an automated object detection and classification. For all scanning tasks, a Metafer Scanning System (Zeiss Axio Imager.Z2 microscope, automatic slide feeder SFx80 and automated oil disperser) from MetaSystems (Altlussheim, GER) was used.
To generate training data for AOI detection, 37 bone marrow smears were scanned at 10x magnification. 6 different quality classes of regions (based on number and distribution of cells) were annotated by hem experts using polygons. In total, 185,000 grid images were extracted from the annotated regions and used for training a deep neural network (DNN) to distinguish the 6 quality classes and to generate a position list for a high resolution scan (40x magnification).
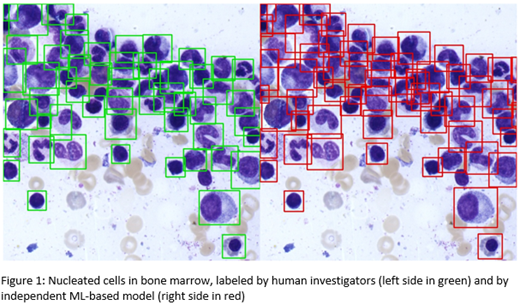
In addition, we scanned the labeled AOI of 68 smears at 40x magnification, acquiring colour images (2048x1496 pixels) of bone marrow cell layers. Each single cell was labeled by human investigators using rectangular bounding boxes (in total: 47,118 cells in 511 images). We set up a supervised ML model, using the labeled 40x images as an input. We fine-tuned the COCO dataset pre-trained YOLOv5 model with our dataset and evaluated using 5-fold cross valuation. To reduce overfitting, image augmentation algorithms were applied.
Results: Our first DNN was able to detect (10x magnification) and capture (40x magnification) AOI in bone marrow smears, sorted by quality and in acceptable time spans. Average time for the 10x pre-scan was 6 min. From the resulting position list, the 50 positions with highest quality values were acquired at an average of 1:30 min.
Our second, independent DNN was able to detect nucleated cells at 94% sensitivity and 75% precision in unlabeled bone marrow images (40x magnification). In this model, we overweighted recall over precision (5:1) to avoid missing any objects of interest, assuming that false positive labels could be corrected by human investigators when reviewing digital images.
For the classification of single cells, a third independent DNN will be necessary. Actually, different approaches are being tested, including our existing blood cell classifier and a former collaborative bone marrow classification model based on a training set of 100,000 annotated bone marrow cells. Depending on these results, new training data for generation of a completely new model could be assessed.
The two existing models enable a fully automated digital workflow including scan of bone marrow smears and delivery of single cell image galleries for human classification already now.
Conclusion: We here present solutions for multiple-DNN-based tools for bone marrow cytomorphology. They allow working digitally and remotely in routine diagnostics. Final solutions will offer single cell classifications and galleries for human review and include real time training of respective classifier models with dynamic datasets.
Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership. Kern: MLL Munich Leukemia Laboratory: Other: Part ownership. Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal